pipeline的构建
1. Pipeline基本概念与规则
Pipeline是Scikit-learn中的一个重要工具,它可以将多个数据处理和模型训练步骤串联在一起。构建Pipeline需要遵循以下规则:
- Pipeline由(名字, 估计器)对的列表构成
- 除最后一个步骤外,其他步骤必须是转换器(transformer)
- 转换器必须具有fit_transform()方法
- 最后一个步骤可以是估计器(estimator)
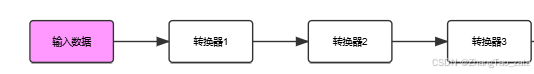
2. Pipeline运行机制
- 执行流程:
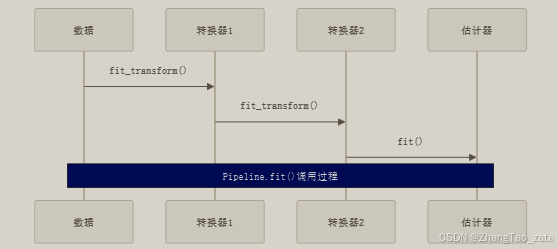
- 关键代码示例:
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.impute import SimpleImputer
# 基础Pipeline构建
num_pipeline = Pipeline([
('imputer', SimpleImputer(strategy="median")),
('std_scaler', StandardScaler()),
])
# 执行转换
data_num_tr = num_pipeline.fit_transform(data_num)
3. 自定义转换器
为了处理Pandas DataFrame,我们需要创建自定义转换器:
from sklearn.base import BaseEstimator, TransformerMixin
class DataFrameSelector(BaseEstimator, TransformerMixin):
def __init__(self, attribute_names):
self.attribute_names = attribute_names
def fit(self, X, y=None):
return self
def transform(self, X):
return X[self.attribute_names].values
4. 完整Pipeline构建流程
- 数值和类别特征处理Pipeline:
graph TD
A[原始数据] --> B[数值特征Pipeline]
A --> C[类别特征Pipeline]
B --> D[特征合并FeatureUnion]
C --> D
D --> E[最终处理后的数据]
subgraph 数值特征处理
B1[DataFrameSelector] --> B2[SimpleImputer]
B2 --> B3[StandardScaler]
end
subgraph 类别特征处理
C1[DataFrameSelector] --> C2[OneHotEncoder]
end
style A fill:#f9f,stroke:#333,stroke-width:2px
style E fill:#bbf,stroke:#333,stroke-width:2px
- 实现代码:
# 数值特征Pipeline
num_pipeline = Pipeline([
('selector', DataFrameSelector(num_attribs)),
('imputer', SimpleImputer(strategy="median")),
('std_scaler', StandardScaler())
])
# 类别特征Pipeline
cat_pipeline = Pipeline([
('selector', DataFrameSelector(cat_attribs)),
('onehot', OneHotEncoder(handle_unknown='ignore')),
])
# 合并Pipeline
from sklearn.pipeline import FeatureUnion
full_pipeline = FeatureUnion(transformer_list=[
('num_pipeline', num_pipeline),
('cat_pipeline', cat_pipeline),
])
5. 使用ColumnTransformer简化Pipeline构建
在Scikit-Learn 0.20及以上版本中,可以使用ColumnTransformer来简化Pipeline构建:
from sklearn.compose import ColumnTransformer
full_pipeline = ColumnTransformer([
("num", num_pipeline, num_attribs),
("cat", OneHotEncoder(), cat_attribs),
])
6. Pipeline的优势
- 代码组织: 将所有预处理步骤组织在一起,使代码更清晰
- 避免数据泄露: 确保在训练集上fit的转换器用相同的参数转换测试集
- 简化工作流: 只需要调用一个fit和transform方法
- 参数优化: 可以对整个pipeline进行网格搜索调优
- 重用性: Pipeline可以保存并重复使用
7. 最佳实践建议
- 为Pipeline中的每个步骤选择有意义的名字
- 确保所有转换器都实现了fit_transform方法
- 使用ColumnTransformer处理不同类型的特征
- 在构建Pipeline之前先测试单个转换器
- 使用get_params()和set_params()方法调整Pipeline参数
- 示例代码
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
import numpy as np
import scipy.linalg
from sklearn.preprocessing import LabelEncoder, StandardScaler
import optuna
import scipy.linalg
from sklearn.linear_model import BayesianRidge
import pandas as pd
from sklearn.model_selection import LeaveOneOut, cross_val_score
class EmscScaler(object):
def __init__(self, order=1):
self.order = order
self._mx = None
def mlr(self, x, y):
"""Multiple linear regression fit of the columns of matrix x
(dependent variables) to constituent vector y (independent variables)
order - order of a smoothing polynomial, which can be included
in the set of independent variables. If order is
not specified, no background will be included.
b - fit coeffs
f - fit result (m x 1 column vector)
r - residual (m x 1 column vector)
"""
if self.order > 0:
s = np.ones((len(y), 1))
for j in range(self.order):
s = np.concatenate((s, (np.arange(0, 1 + (1.0 / (len(y) - 1)), 1.0 / (len(y) - 1)) ** j).reshape(-1,1)[0:len(y)]),1)
X = np.concatenate((x.reshape(-1,1), s), 1)
else:
X = x
# calc fit b=fit coefficients
b = np.dot(np.dot(scipy.linalg.pinv(np.dot(X.T, X)), X.T), y)
f = np.dot(X, b)
r = y - f
return b, f, r
def fit(self, X, y=None):
"""fit to X (get average spectrum), y is a passthrough for pipeline compatibility"""
self._mx = np.mean(X, axis=0)
def transform(self, X, y=None, copy=None):
if type(self._mx) == type(None):
print("EMSC not fit yet. run .fit method on reference spectra")
else:
# do fitting
corr = np.zeros(X.shape)
for i in range(len(X)):
b, f, r = self.mlr(self._mx, X[i, :])
corr[i, :] = np.reshape((r / b[0]) + self._mx, (corr.shape[1],))
return corr
def fit_transform(self, X, y=None):
self.fit(X)
return self.transform(X)
from sklearn.base import BaseEstimator, TransformerMixin
class SpectraPreprocessor(BaseEstimator, TransformerMixin):
def __init__(self, emsc_order=3,X_ref=None):
self.emsc_order = emsc_order
self.emsc_scalers = [EmscScaler(order=emsc_order) for _ in range(4)]
self.X_ref = X_ref
def fit(self, X, y=None):
X_ref = self.X_ref
if X_ref is None:
X_ref = X.copy()
# Define the column ranges for each segment
ranges = [(0, 251), (281, 482), (482, 683), (683, 854)]
# Fit EmscScaler for each segment
for i, (start, end) in enumerate(ranges):
self.emsc_scalers[i].fit(X_ref[:, start:end])
return self
def transform(self, X, y=None):
# Define the column ranges for each segment
ranges = [(0, 251), (281, 482), (482, 683), (683, 854)]
# Transform each segment
transformed_segments = []
for i, (start, end) in enumerate(ranges):
segment = X[:, start:end]
transformed_segment = self.emsc_scalers[i].transform(segment)
transformed_segments.append(transformed_segment)
# Concatenate all transformed segments
return np.concatenate(transformed_segments, axis=1)
def fit_transform(self, X, y=None):
self.fit(X)
return self.transform(X)
def bayesian_ridge_optuna_for_emsc_data(x_train, y_train, pipeline_):
def objective(trial):
try:
alpha_1 = trial.suggest_float('alpha_1', 0.001, 1, log=True)
alpha_2 = trial.suggest_float('alpha_2', 0.001, 1, log=True)
lambda_1 = trial.suggest_float('lambda_1', 0.001, 1, log=True)
lambda_2 = trial.suggest_float('lambda_2', 0.001, 1, log=True)
model = pipeline_.set_params(
bayesian_ridge__alpha_1=alpha_1,
bayesian_ridge__alpha_2=alpha_2,
bayesian_ridge__lambda_1=lambda_1,
bayesian_ridge__lambda_2=lambda_2
)
model.fit(x_train, y_train)
score = cross_val_score(model, x_train, y_train, cv=10, n_jobs=-1, scoring='r2')
return np.mean(score)
except ValueError as e:
return -np.inf
optuna.logging.set_verbosity(optuna.logging.WARNING)
pruner = optuna.pruners.MedianPruner()
study = optuna.create_study(direction="maximize", pruner=pruner)
study.optimize(objective, n_trials=500, show_progress_bar=True, n_jobs=1)
return study.best_params
def getdata(filenamex, filenamey):
x = pd.read_csv(filenamex, header=None)
y = pd.read_csv(filenamey)
data = pd.concat([x, y], axis=1)
return data
name = 'test'
x, y = np.random.rand(100,884), np.random.rand(100)
x_ref = np.random.rand(30,884)
pipeline = Pipeline([
('preprocessor', SpectraPreprocessor(emsc_order=3, X_ref=None)),
('scaler', StandardScaler()),
('bayesian_ridge', BayesianRidge())
])
pipeline.set_params(preprocessor__X_ref=x_ref)
############################################################################################################################################################
best_params = bayesian_ridge_optuna_for_emsc_data(x, y, pipeline)
############################################################################################################################################################
pipeline.set_params(
bayesian_ridge__alpha_1=best_params['alpha_1'],
bayesian_ridge__alpha_2=best_params['alpha_2'],
bayesian_ridge__lambda_1=best_params['lambda_1'],
bayesian_ridge__lambda_2=best_params['lambda_2']
)
pipeline.fit(x, y)
y_pred = pipeline.predict(x)
print(y_pred)
